Overview of Single-Cell Sequencing: Isolation Methods and Techniques
Bulk RNA-seq has been used since the late 2000s as a tool to study the whole transcriptome. However, the data generated with this approach represents an average of the gene expression within a sample. To bring higher resolution to this experimental approach, single-cell RNA-sequencing (scRNA-seq) was developed to provide the same utility of transcriptomic characterization to individual cells. scRNA-seq is becoming more and more common in biological studies as it provides an unprecedented view of biological changes within specific cell subtypes in a heterogenous population.
Though scRNA-seq is becoming more prevalent, the exact methods and terminology used to describe these techniques are constantly evolving and can be hard to differentiate. Bulk and single-cell sequencing approaches share similar library preparation and sequencing steps, but are very different in terms of sample preparation, particularly in methods used to isolate single cells. Here, we’ll provide an overview and comparison of commonly used single-cell isolation methods.
Single-Cell Sorting and Microfluidics
One of the earliest methods to sort single cells was using limiting dilutions or single-cell fluorescence-activated cell sorting (FACS). These approaches are lower throughput as there is a limit to the number of cells that can be analyzed before the experiment becomes too difficult to manage.
Another slightly higher-throughput method involves using microfluidic systems like the Fluidigm C1™ and Takara ICell8 nanowell chips. In this nanowell-based method, wells are loaded by passing a suspension of cells over the wells. The wells are often examined by microscopy to ensure they contain single cells before continuing. Once in the wells, the samples are prepared using a technique like Smart-seq21,2, which independently isolates the RNA of each cell. It is essentially the same as a bulk RNA-seq experiment, except that the input is a single cell instead of a larger sample. These techniques are advantageous because they capture all RNA, not just mRNA, and the efficiency of measuring many transcripts is very high compared to other methods. However, they are limited by the number of cells that can analyzed, and the process can be labor intensive.
Bead-Based Methods
Microfluidic chips can be utilized to form nanoliter-sized aquaeous droplets in oil that isolates single cells. Within these droplets, the individual cells are encapsulated with a bead that is coated in poly(dT) oligonucleotides to capture mRNA. These oligonucleotides also contain a cell barcode unique to each bead to identify the cell and a unique molecular identifier (UMI), which is used to identify each unique transcript. Once the captured mRNA is reverse transcribed, the cell barcode and UMI are incorporated
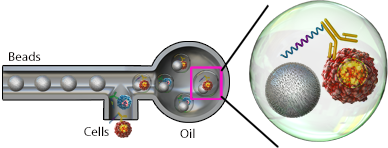
Example of bead-based droplet encapsulation.
into each cDNA molecule. These cDNAs can then be pooled for sequencing where each gene is counted by the UMI and assigned to a cell by the cell barcode. This results in a very high-throughput method to interrogate tens of thousands of cells; however, the efficiency of RNA capture is lower than techniques that are able to capture all RNA, like nanowell chips.
Many iterations of this technology have been developed, with the earliest ones being homebrewed by individual labs, like Drop-seq3 and inDrop4. Commercial systems based on droplet partition have been developed by several companies since then, including 10x Genomics, BioRad, Dolomite Bio, and 1CellBio. In addition, Mission Bio’s platform obtains single cell genomic information by enriching genes of interest with panels.
Finally, there are bead-based nanowell techniques. These nanowells contain an oligonucleotide-conjugated bead, similar to the droplet-based approach. A single cell and oligo-conjugated bead are loaded into each well and reagents are added to perform reverse transcription. From there, the cDNA is pooled and prepared for sequencing. Unlike the Smart-seq methods that isolate total RNA from the cell, the mRNA is being captured by a bead in this approach. This technique has been commercialized by CelSee (now part of BioRad) and Becton Dickinson. Newer applications, like CITE-seq and REAP-seq, add protein detection by sequencing to these workflows. Cells are first stained with oligo-conjugated antibodies, like our TotalSeq™ reagents, containing a capture sequence to bind to the bead, a second barcode that is specific to the antibody to identify the protein, and a PCR handle for sequencing. We offer a variety formats of our oligo-conjugated reagents to integrate into multiple droplet-based single-cell isolation platforms.
Regardless of how the cells were isolated, once the single-cell libraries are prepared, they are sequenced, typically using next generation sequencing (NGS) with an Illumina instrument. The libraries are then aligned to the genome and assigned to a cell barcode, if required. The downstream analysis requires bioinformatics tools, which we will be discussing in a future blog. The development of single-cell sequencing technology has propelled biology into a new era of discovery. Having the ability to analyze tissue at the most basic biological unit, a cell, can resolve many questions that, until now, have remained unanswered.
| Summary of Single-Cell Isolation Methods | |||
|---|---|---|---|
| Method | Benefits | Limitations | Throughput |
| Limiting Dilutions or FACS | Isolates total RNA and measures many transcripts with high efficiency Doesn’t require specialized equipment |
Low-throughput, limited by the number of number of cells that can be analyzed Process can be labor intensive |
Low |
| Nanowell Chips | Isolates total RNA and measures many transcripts with high efficiency | Medium-throughput compared to bead-based applications | Medium |
| Droplet-Based Methods | Very high-throughput, can investigate tens of thousands of cells | Only captures a subset of mRNAs | High |
| Bead-Based Nanowell Applications | Very high-throughput Easily scalable for even larger cell numbers |
Only captures a subset of mRNAs | High |
References:
- Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
- Picelli, S. et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013).
- Macosko, E. Z. et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214 (2015).
- Klein, A. M. et al. Droplet barcoding for single cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015).






Follow Us