SARS-CoV-2 is the virus that causes COVID-19, a respiratory disease responsible for the worldwide pandemic that started in 2019. SARS-CoV-2 belongs to the Coronaviridae family, which also includes SARS-CoV and MERS-CoV, all of which are enveloped, single-stranded, positive sense RNA viruses1. Understanding the functions of each protein encoded by the viral genome is critical to dismantling the biology and pathology of COVID-19. Here we cover the functions of key SARS-CoV-2 proteins and the high-quality research reagents BioLegend offers to support research on these proteins.
SARS-CoV-2 Proteins
Virion Structure and Assembly
SARS-CoV-2 encodes 16 nonstructural proteins, 9 accessory proteins, and 4 main structural proteins: spike protein (S), membrane protein (M), envelope protein (E), and nucleocapsid protein (N). Infected cells produce viral proteins that are expressed in the ER and Golgi complex, where they assemble with viral RNA. The virus uses internal host cell membranes to form individual virions by budding into intracellular vesicles. Viruses are subsequently released from cells via exocytosis as 70-90 nm diameter particles.
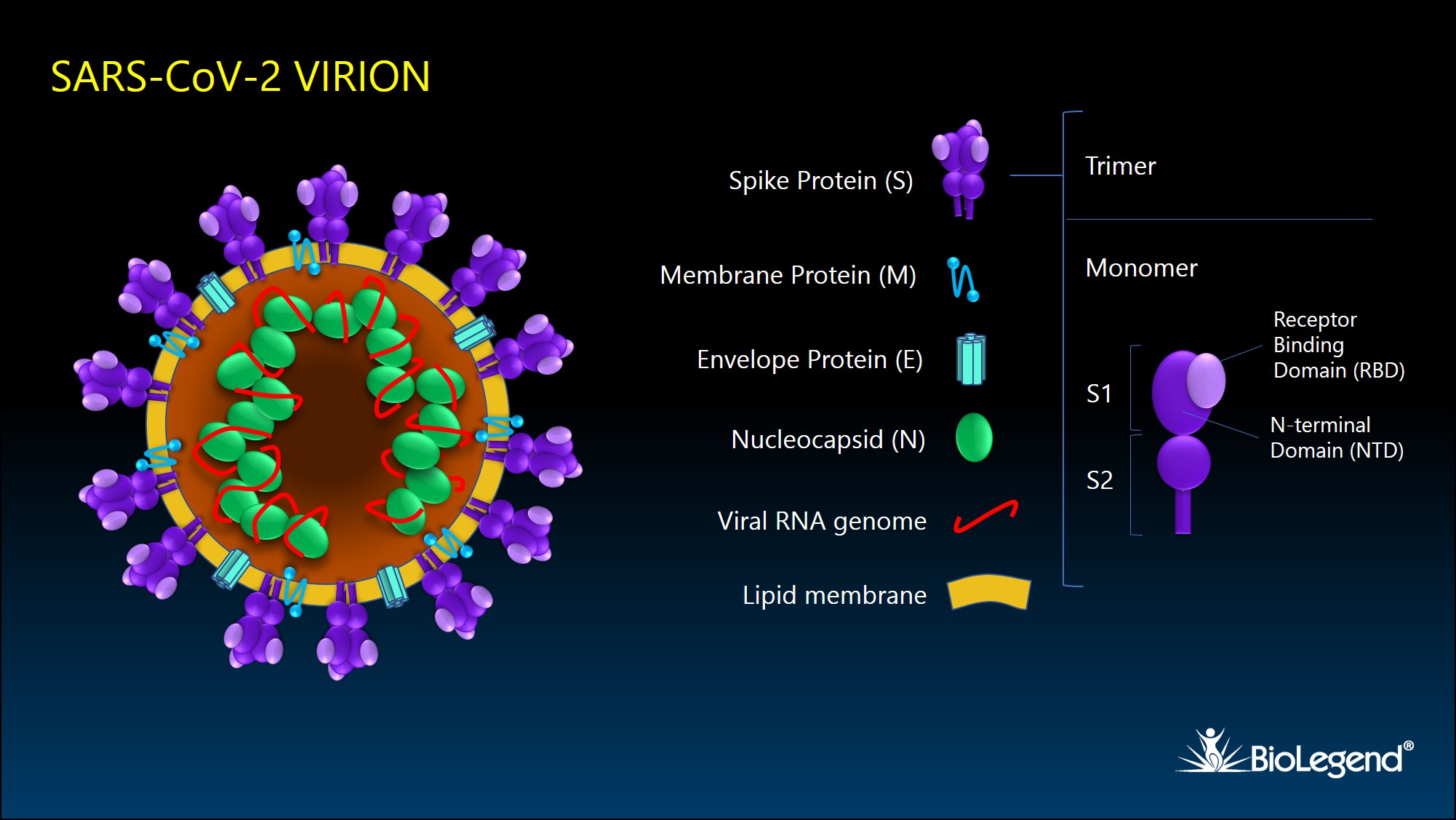
Figure 1. Schematic illustration of SARS-CoV-2 viral particle and its structural proteins.
Spike Protein (S) and Variants
The SARS-CoV-2 spike glycoprotein, or S protein, binds to target receptors on the host cell surface. It is synthesized as a 1300 aa precursor protein that is cleaved into an N-terminal S1 subunit and a C-terminal S2 subunit. The S1 subunit is comprised of 2 domains, the receptor binding domain (RBD) and the N-terminal domain (NTD). Three S1/S2 heterodimers assemble to form a trimer spike protruding from the viral envelope. Binding of the RBD with host surface receptor protein, ACE2, initiates membrane fusion, which is mediated by a FP (fusion peptide) domain and two heptad repeat domains (HR1 and HR2) of the S2 subunit2.
The spike protein serves as the target for first generation vaccines, which have demonstrated efficacy in neutralizing the virus and preventing spread. This initial wild-type sequence (Genbank accession: QHD43416) was published in January of 2020. During adaptations to new human hosts, mutations have led to variants that are either more virulent, more transmissible, or both3. The Delta variant (B.1.617.2) emerged in the US during summer of 2021 and has led to rebounding numbers of cases and hospitalizations.
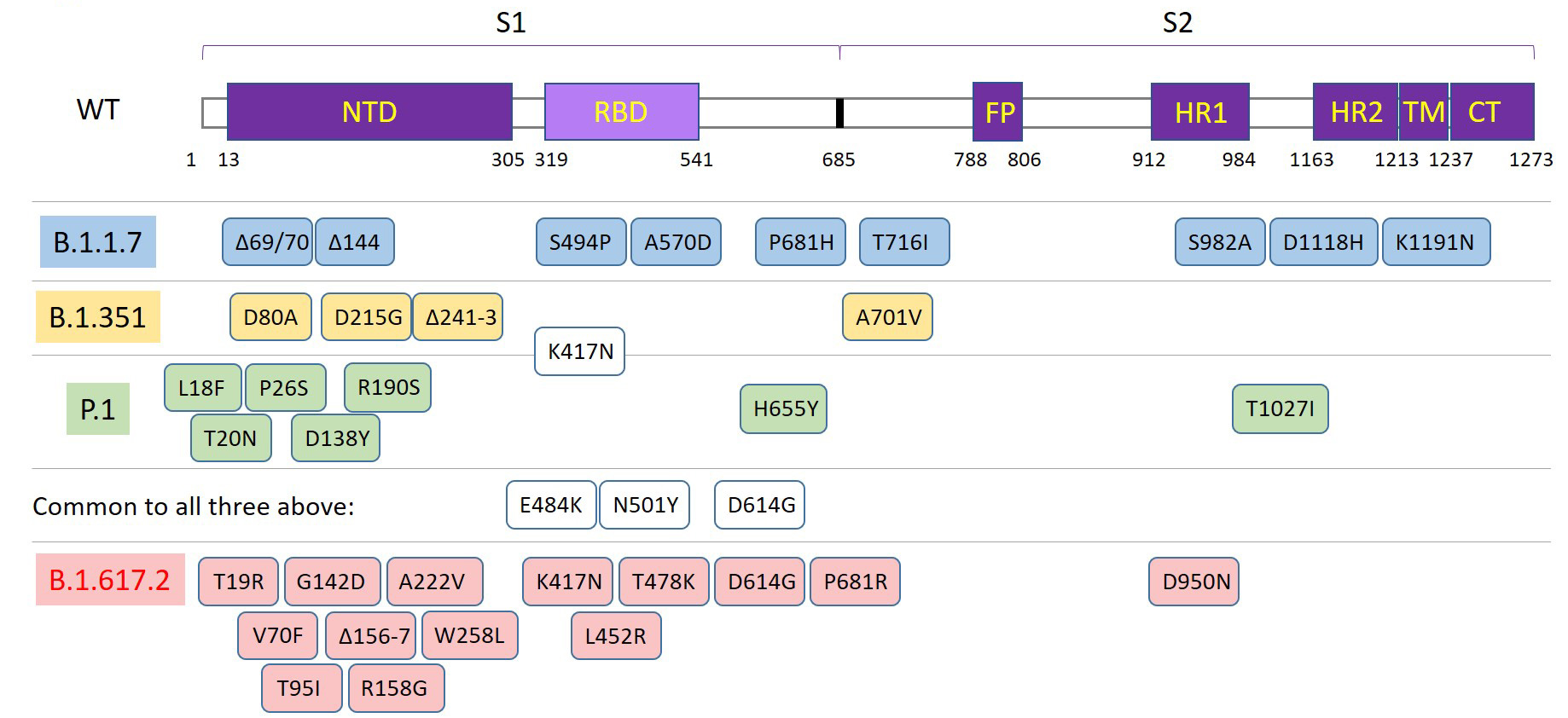
Figure 2. Diagram of the spike protein domains and mutations associated with the major variants at this time.
SARS-CoV-2 Proteins and Reagents
Spike Protein (S)
BioLegend offers an assortment of research reagents specific to the spike protein for the study of SARS-CoV-2, including antibodies and recombinant proteins. Learn more about our full reagent sets for the spike protein below.
Recombinant Proteins
Each lot of every recombinant protein product is quality-tested for binding to recombinant ACE2, or other biologically relevant bioassays. One critical reagent is our furin-resistant recombinant Spike S1+S2 that enables research applications where the intact protein is desired, such as receptor binding assays or cell binding assays.
Primary Antibodies
Our highly specific anti-spike protein antibodies are suitable and quality-tested for a variety of applications: Western blots, ELISA, flow cytometry, or bioassays like the blocking of receptor/virus interaction. Find the best suited antibody for your applications below. Click the icons to view products.
S1
View products for:
Blocking
Western Blotting
Flow Cytometry
Direct ELISA
S2
View products for:
Western Blotting
Direct ELISA
RBD
View products for:
Blocking
Membrane Protein (M)
The SARS-CoV-2 membrane glycoprotein (M) is vital for assembly and budding of new viral particles. By organizing and assisting in the assembly of viral structural proteins, it is pivotal in dictating the size and shape of virions. It directly interacts with spike (S), envelope (E), and nucleocapsid (N) proteins, as well as other non-structural proteins. Additionally, it may play a role in viral transmission by assisting S protein during cell attachment and entry.
Envelope Protein (E)
The envelope protein (E) forms pentamers that serve as ion channels, referred to as viroporins. The protein is known to play a role in the pathogenicity of the virus, while also playing a role in assembly and release of virions. It activates host cell NLRP3 inflammasome pathways, leading to overexpression of IL-1β. Interestingly, it also bears a PDZ-binding motif that mediates interaction with the host protein syntenin, which activates MAPK signaling and downstream release of inflammatory cytokines. Both pathways can lead to activation of cytokine storm and related pathologies4.
Nucleocapsid Protein (N)
The SARS-CoV-2 nucleocapsid (N) protein directly interacts with the viral genome and helps to package it into newly forming virions. The protein forms dimers and binds to RNA in a sequence non-specific manner. The SARS-CoV-2 nucleocapsid protein consists of five domains: an N-terminal tail region, an N-terminal RNA binding domain (N-NTD), a Ser/Arg-rich linker region (LKR), a C-terminal dimerization domain (N-CTD), and a C-terminal intrinsically disordered region (IDR)5. We provide tools for the quantitative measurement of nucleocapsid protein.
Non-structural proteins (Nsps)
Non-structural proteins constitute approximately half of the SARS-CoV-2 genome, encoding 16 proteins. While some protein functions remain unknown, the majority play a role in the viral life cycle, including formation of replication-transcription complexes, suppressing host gene expression, enzymatic cleavage of mature and intermediate Nsps, inducing double membrane vesicles, RNA polymerase activity, exoribonuclease and endoribonuclease activity, and methyltransferase activity6. Nsp5 of SARS-CoV-2, also known as 3C-like protease (3CLpro) or main protease (Mpro), cleaves the two main polyproteins, pp1a and pp1ab, at 11 cleavage sites, helping produce the 16 individual Nsps7. Not surprisingly, it is considered an important target for inhibiting the viral life cycle.
View products related to SARS-CoV-2 Nsps
Accessory Proteins
SARS-CoV-2 accessory proteins are encoded by 9 open reading frames (Orf) and are named Orf3a, Orf3b, Orf6, Orf7a, Orf7b, Orf8, Orf9b, Orf9c, and Orf10. These proteins are the least understood of all the viral proteins, but some have clearly defined functions6. Orf3a interacts with structural proteins N, M, and S, and serves as an ion channel that likely contributes to virus budding. Orf9b is an E3 ubiquitin ligase that has been shown to interfere with the cellular machinery for the induction of IFN responses, while also possibly being involved in virus assembly. Orf3b, Orf6, Orf8, Orf9b, and Orf10 may have similar inhibitory functions on antiviral host signaling. Orf7a may serve to support virus budding and release. Orf8 binds to MHC-I, resulting in decreased antigen presentation.
A Full Suite of Tools for Dismantling SARS-CoV-2
Whether you are doing basic research, developing vaccines, or analyzing patient samples, BioLegend provides quality reagents for your studies. Learn more.
BioLegend now offers a multiomics method to characterize antigen-specific B cell mediated responses in COVID-19 and other infectious diseases. Check out LIBRA-seq.
References
- Hu B., et al. 2021. Nat Rev Microbiol. 19:141. Pubmed
- Huang Y., et al. 2020. Acta Pharmacol Sin. 41:1141. Pubmed
- https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html
- Schoeman D. and Fielding B. 2020. Front Microbiol. 11:2086. Pubmed
- Yang M., et al. 2021. Front Chem. 8:624765. Pubmed
- Mariano G., et al. 2020. Front Mol Biosci. 7:605236. Pubmed
- Rathnayake A.D., et al. 2020. Sci Transl Med. 12:eabc5332. Pubmed
 Login / Register
Login / Register 






Follow Us