Our Solutions for Characterizing the B cell Mediated Response
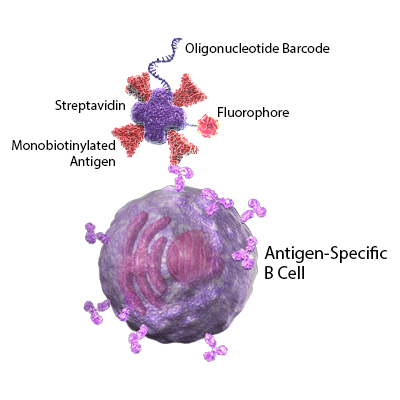
We provide a ready-to-use system for labeling SARS-CoV-2 antigens with oligonucleotides and fluorophores, removing the need for lengthy workflows associated with synthesizing and conjugating complex molecules to these antigens.
For more information or guidance on how to use these products, contact our technical services team.
Monobiotinylated Recombinant Proteins

The SARS-CoV-2 spike glycoprotein is synthesized as a precursor protein that can be cleaved into an N-terminal S1 subunit and a C-terminal S2 subunit. Binding of the spike protein with host surface proteins initiates membrane fusion and viral entry. Leveraging our recombinant protein expression and modification expertise, we provide high-quality monobiotinylated S protein RBD and S1 subunits.
Biotinylated Recombinant SARS-CoV-2 S Protein RBD Biotinylated Recombinant SARS-CoV-2 S Protein S1
TotalSeq™-C Streptavidin Conjugates

Our TotalSeq™ oligonucleotide and fluorophore conjugated streptavidin allows for the multimerization of SARS-CoV-2 antigens, provides a unique antigen barcode, and enables fluorescent-based cell sorting prior to sequencing. Because our TotalSeq™-C products seamlessly integrate with the 10x Genomics Immune Profiling solution for BCR sequencing and RNA characterization, there is no need to validate oligos for overlap with 10x Genomics sequences.
View all TotalSeq™-C Streptavidin Reagents
Additional Related Products
TotalSeq™ oligo-conjugated reagents enable protein detection by sequencing and integrate seamlessly into existing single-cell transcriptomics, bulk sequencing, or single-cell genomics workflows.
Learn more about TotalSeq™ Reagents
 Login/Register
Login/Register 







Follow Us