Uncovering Immune Signatures With TotalSeq™ Universal Cocktails
Single-cell multiomics expands protein detection beyond traditional flow cytometry techniques, permitting the detection of >100 antibodies in a single assay. Because this application relies on oligonucleotide sequencing as a readout, not fluorophores which have a discrete excitation and emission profile, hundreds of proteins can be detected in a single experiment.
Simultaneous detection of this many antibodies, however, comes with some technical considerations. Each antibody should be properly titrated to ensure balanced read counts. To make this process easier, our scientists developed TotalSeq™ Universal Cocktails. Each cocktail contains over 115 antibodies and their associated isotype controls for large-scale protein detection. Within the cocktail, the antibodies are individually titrated using next-generation sequencing to provide optimal discrimination of positive and negative cell populations.
Our antibody cocktails have been a part of important breakthroughs including discovering risk factors of long COVID, identifying immune cell signatures of pediatric multisystem inflammatory disease, and creating a longitudinal single-cell atlas of COVID-19. In this blog, we highlight recent research demonstrating how researchers have used our TotalSeq™ cocktails in top journals to study the impact and pathophysiology of COVID-19.
Watch our on-demand webinar with Dr. Aaron Streets as he uses single-cell multiomics and an early version of our TotalSeq™ mouse universal cocktail to study thymocyte development.
Research Highlights
Sacco et al. Nature Medicine. Feb 2022
While COVID-19 has not been severe in most pediatric patients, some children that have been infected with SARS-CoV-2 develop a hyperinflammatory response known as multisystem inflammatory syndrome (MIS-C). This Nature Medicine longitudinal study used multiomics analysis to perform immunological profiling of children with COVID-19, those that developed MIS-C, and healthy controls. The authors used a large TotalSeq™-C panel to perform CITE-seq, correlating the expression of 188 proteins with transcriptome analysis and the immune repertoire.
The authors were able to identify immune signatures unique to pediatric COVID-19 patients and those that develop MIS-C. Importantly, COVID-19 patients showed a strong type I interferon response, while patients that developed MIS-C exhibited type II IFN-dependent and NF-κB-dependent signatures and had increased levels of circulating spike protein. There was an association of MIS-C with specific T cell clonotypes and certain HLA alleles, which suggested that there may be genetic susceptibilities. Lastly, MIS-C B cells showed a higher mutational load. Together, these large datasets form a pathophysiological signature that can inform treatment options and help determine the cause of MIS-C.
Multiple early factors anticipate post-acute COVID-19 sequelae
Su Y. et al. Cell. March 2022.
The risk factors and treatment of post-acute sequelae of COVID-19 (PASC), often referred to as “long COVID,” are incompletely defined, in part, due to the heterogeneity of the condition and the diverse factors that may contribute. A recent study in Cell used a systems-based, longitudinal multiomics approach to follow COVID-19 patients and identify factors associated with PASC. As part of their study, they performed single-cell CITE-seq using a large, custom TotalSeq™-C antibody panel to study the expression of 192 cell surface proteins.
Ultimately, they identified four major factors that correlate with the development of PASC including pre-existing diabetes, high levels of SARS-CoV-2 RNA in the blood, reactivation of latent Epstein-Barr virus (EBV), and the presence of specific autoantibodies. This study provides a framework for further research to better resolve how these factors contribute to PASC and can guide the development of treatments.
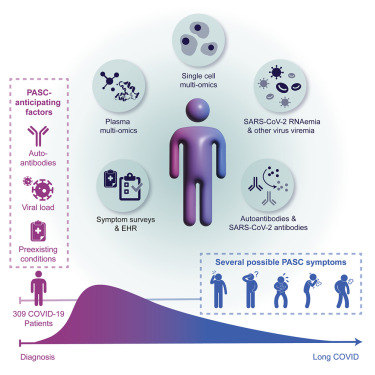
Graphical abstract from Su Y, et al. Cell. 185: 885-895. CC BY 4.0.
Time-resolved systems immunology reveals a late juncture linked to fatal COVID-19
Liu C. et al. Cell. April 2021.
Creating a single-cell atlas from COVID-19 patients, this study in Cell used a longitudinal assessment of circulating proteins and 188 surface protein markers, transcriptome, and T cell receptor sequence simultaneously in PBMCs. By performing a longitudinal study, they were able to link immune response variation to disease severity over time. Using our TotalSeq™ hashtag reagents and a large cell TotalSeq™ antibody cocktail, they used single-cell surface protein expression to identify 30 immune cell subsets and track gene expression changes within these subsets.
Ultimately, their study revealed diverging inflammatory and host immune responses that predicted patient recovery. A longitudinal analysis allowed for the classification of innate and adaptive immune cell types and subsets over time providing a refined look at individual clusters of lineage-specific cell identification. These datasets revealed that disease severity correlated with decreased plasmacytoid dendritic cell frequency and NK cell dysfunction that is linked to IL-15 and decreased immune signatures. These changes presented at a late juncture in COVID-19 infection.
The addition of oligo-conjugated antibodies to large single-cell experiments can reveal new insights by combining protein data with transcriptome measurements and TCR/BCR profiling. Using these data, we can determine the immune signature of patients at varying stages of diseases or with different states of disease severity. Large-scale protein detection can be technically challenging to adopt; however, our TotalSeq™ universal cocktails provide a ready-to-use solution to incorporate large-scale protein detection into your single-cell experiments.
These longitudinal multiomics approaches advanced our understanding of SARS-CoV-2 infection and how it can develop into long COVID and MIS-C. Using our TotalSeq™️ antibody cocktails for large-scale protein detection, these studies identified unique immune signatures to discover potential risk factors and therapeutic targets.
 Login / Register
Login / Register 






Follow Us