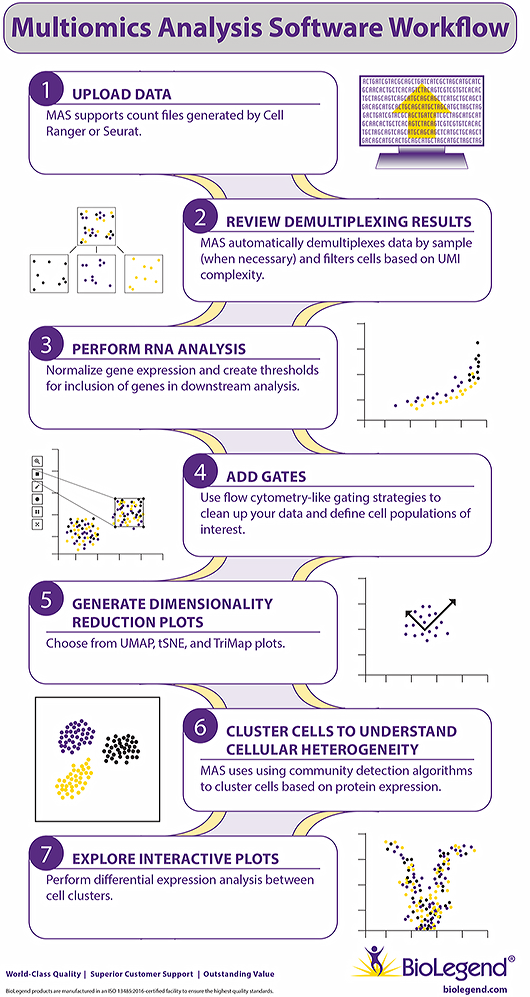
Single-cell proteogenomic analysis often requires advanced tools and experience to process and interrogate the data, which may not be readily available. To make single-cell data analysis easier, we offer Multiomics Analysis Software (MAS). MAS is a free cloud-based program that allows users to quickly and easily explore CITE-seq data and create publication-quality images without extensive bioinformatics knowledge.
Multiomics Analysis Software
MAS Key Features
- A simple user interface for exploring CITE-Seq data.
- Accessible to all users irrespective of bioinformatics background.
- Accessible through your web browser, no server access required.
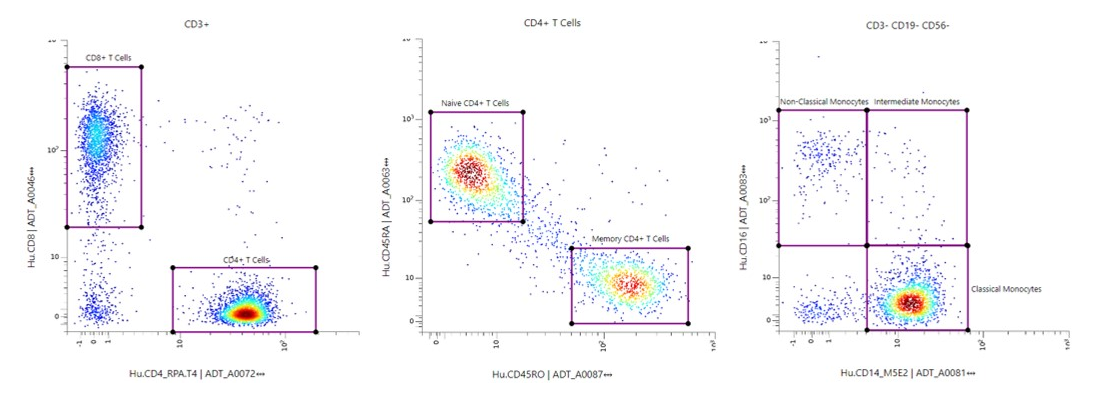
- Uses flow cytometry-like gating based on TotalSeq™ antibody staining.
- Creates UMAP, tSNE, or TriMAP dimensionality reduction plots.
- Easily share any workspace with collaborators or BioLegend Technical Services.
- Exportable in *.h5ad format for use in other single-cell analysis programs.
MAS is a free cloud-based software. Before using the software suite, you will need to request an account. To request an account please fill in the form below. Please note an institutional email address is required. Generic email addresses such as Gmail, iCloud, Yahoo, etc. will not be accepted.
Request an Account
What Can MAS Do?
Use Flow-like Gating to Identify Cells Based on TotalSeq™️ Expression
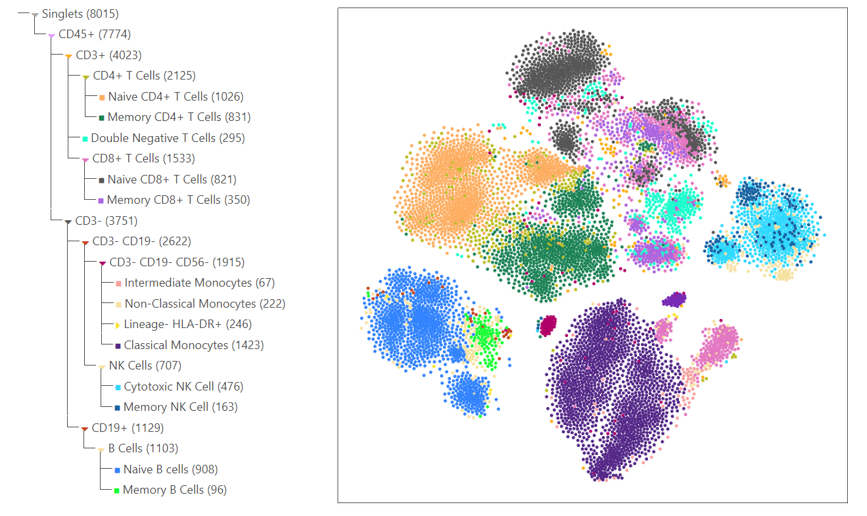
Create Interactive Dimensionality Reduction Plots such as UMAPs and t-SNEs
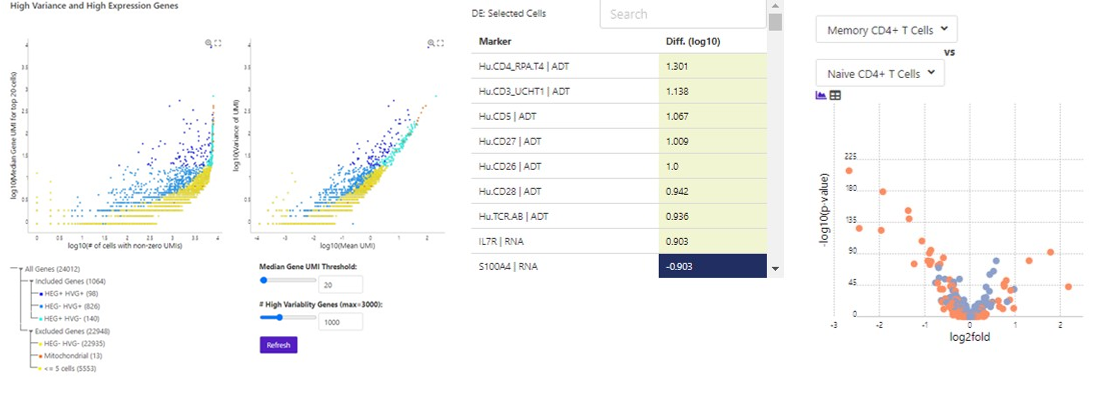
Perform HVG Analysis to Identify Genes
of Interest
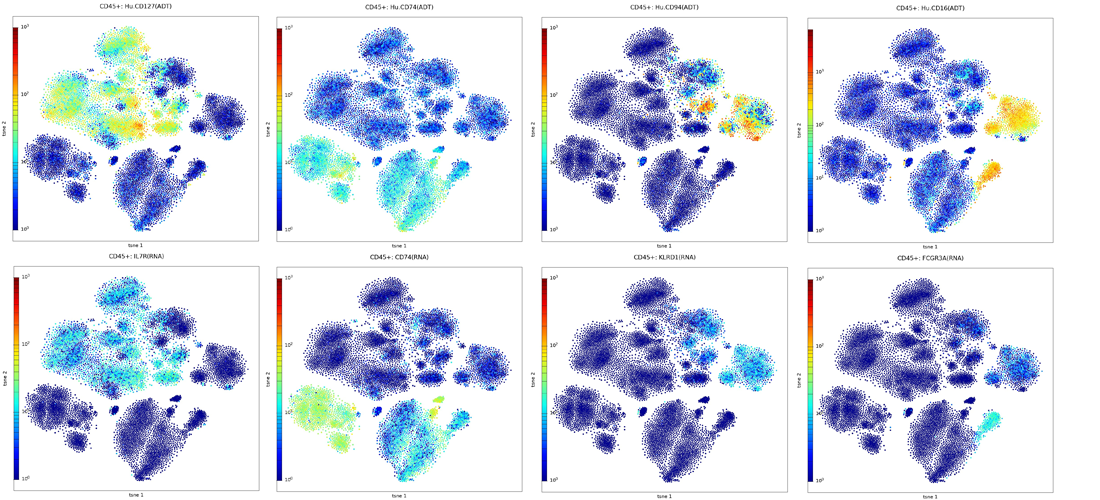
Explore Protein and RNA Expression in
Subpopulations of Cells
Compare Clusters to Identify Markers
of Interest
Visualize Protein and RNA expression
Support Tools for Getting Started with MAS
Sample Data Set
If you’d like to start exploring the software, but don’t have single-cell data to explore, we have sample datasets and a video tutorial below for your convenience. To learn more about Totalseq data analysis and to explore additional example datasets, please check out our data analysis page.
If you have a feature barcode matrix you’d like to explore, request an account under the "Access the Software" tab. Make sure the Features.tsv.gz file adheres to the following guidelines:
- Rows containing cell hashing antibodies, need to have “HTO_###” in column A and “Cell Hashing” listed in column C
- Rows containing non-cell hashing antibodies, need to have “ADT_###” in column A and “Antibody Capture” listed in column C
User Manual
The User Manual contains all essential information for the use of the MAS, including a description of its functions and capabilities, as well as step-by-step procedures for access and use.
 Login / Register
Login / Register 






Follow Us