A Simplified Multiomics Workflow with Pre-Titrated TotalSeq™ Antibody Cocktails
Incorporating antibodies into your multiomics workflow requires optimization to ensure the best results. For example, titrating individual antibodies ensures that you can clearly identify the positive population without using too much of your sequencing depth for any one target.
However, if you’re planning an experiment containing high-plexity antibody staining, the process of titrating and balancing the entire panel can be tedious, time-consuming, and costly. Ideally, you would titrate each antibody by flow cytometry, followed by additional titration and balancing of reads using next-generation sequencing (NGS). Both of these processes require a significant amount of time and money.
Simplify your workflow with our expertly-designed antibody cocktails that remove the need for this additional optimization and minimize variability between experiments. Each cocktail comes as a single-use tube containing pre-titrated amounts of lyophilized antibodies and is quality tested using NGS as a readout.
Explore our TotalSeq™ antibody panels.
Here, we’ll walk you through our process for developing and optimizing each of our antibody panels to help you understand the rigorous methods we use to ensure that our oligo-antibody cocktails provide optimal results and reduce your experimental costs.
Cocktail Conceptualization to Creation
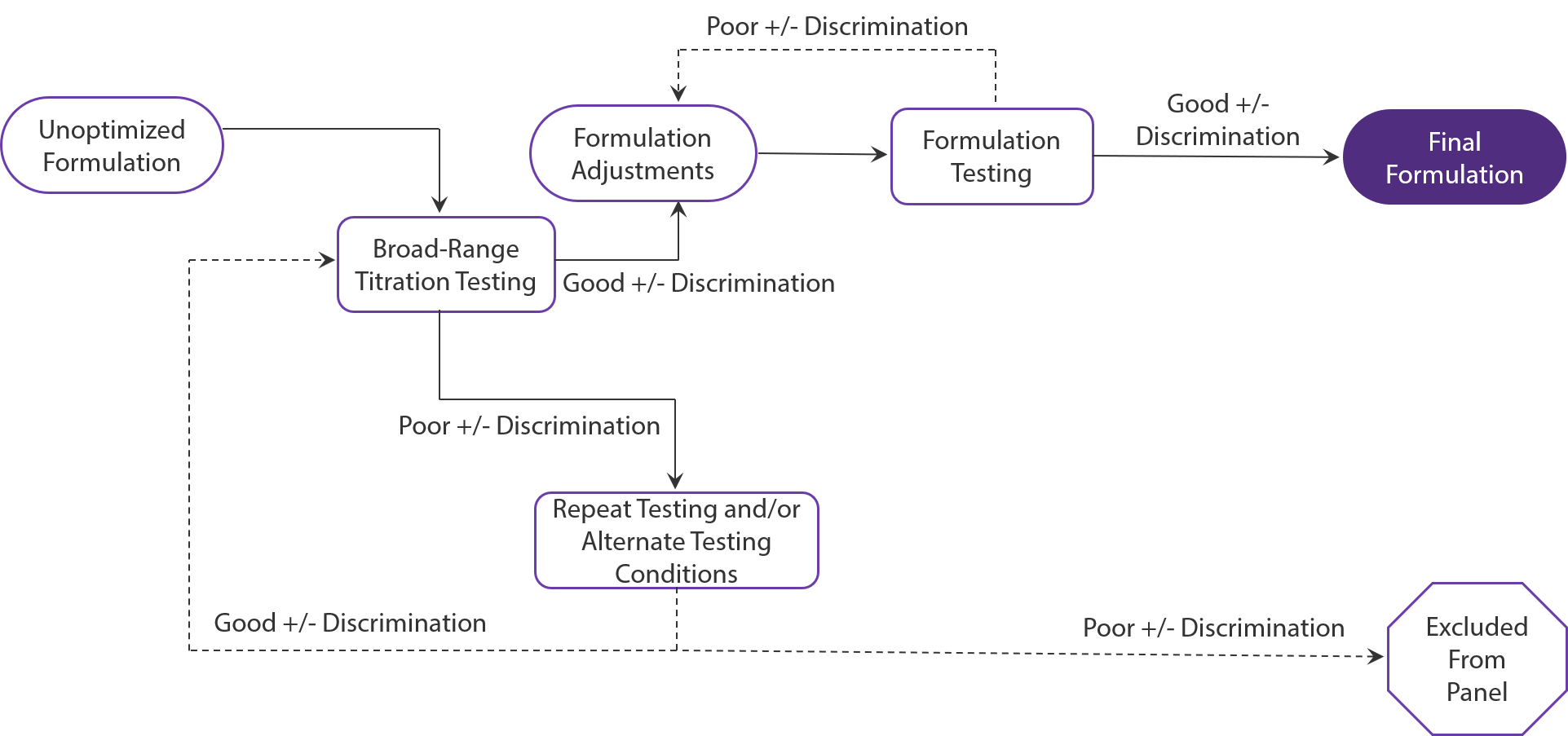
Figure 1: The process of TotalSeq™ antibody cocktail conceptualization, validation, and creation.
The development of our antibody cocktails is an extensive process from conceptualization to creation. Each cocktail first begins as an unoptimized formulation. From there, it undergoes titration testing using primary cells and cell lines under various stimulation conditions. Antibody concentrations that provide clear discrimination between the positive and negative population and distribution of UMI counts are kept in the formulation for further testing.
Antibodies that demonstrate poor discrimination are tested using alternate conditions. If performance cannot be recovered, the antibody is excluded from the cocktail. Once the antibodies have been selected and adjusted, the cocktail undergoes further testing with single-cell experiments which may result in further optimizations as we prepare the final formulation
Antibody Titration
Before release, our scientists fully optimize and titrate each panel. Properly titrating antibodies for use in multiomics applications, like CITE-seq, facilitates read balancing across antibodies, reduces signal-to-noise, and decreases unnecessary sequencing reads. While the process of titrating an individual antibody is fairly straightforward, it becomes more challenging with an increasing number of antibodies in a panel.
Figure 2: TotalSeq™ antibody titration using NGS. The x-axis shows a ranked ordering of cells and the y-axis represents the UMI counts for each antibody.
For a typical titration experiment, we identify a base concentration using flow cytometry staining with the same antibody clone. From there, we perform a serial dilution of the oligo-conjugated antibody, stain cells, generate libraries, and sequence. Figure 2 shows a ranked ordering of cells on the x-axis and the UMI counts for each antibody on the y-axis. The middle of the positive peak represents the mean UMI count for each dilution. In yellow, we’ve highlighted the concentration chosen for each specific antibody in a panel.
The goal for antibody staining in a multiomics experiment is not to obtain maximum separation between the positive and negative peaks, but instead to find the optimal antibody concentration at which there is enough separation to easily discriminate between positive and negative cells.
Balancing Read Distribution
Once individual titrations have been completed, we pool each antibody at the determined concentration and perform further sequencing. This process confirms proper titration and ensures we have a balanced distribution of reads for each antibody across the panel.
Figure 3: The percentage of sequencing reads from each antibody in a multiomics experiment. The left column used an unoptimized concentration of antibody and the right column shows the percentage of reads following titration and optimization.
In figure 3, we show data from two separate titration experiments. On the left column are the results of a staining experiment with an unoptimized concentration of an anti-CD54 TotalSeq™-A antibody. A total of 8.5% of pass-filter sequencing reads are attributed to this one antibody. On the right-hand column, we see the results of a subsequent validation staining experiment with a panel containing an optimized concentration of the anti-CD54 antibody. In this experiment, we’re able to reduce the percentage of sequencing reads attributed to this antibody by approximately 10-fold while retaining the ability to discriminate positive cells. This particular antibody is now balanced with the other antibodies in the panel.
When we build a panel, we find antibody concentrations that achieve the best balance in sequencing reads. If some markers represent a disproportionate fraction of the entire sequencing data, such as CD54 in the example here, this may reduce your ability to identify rarer cell markers and add unnecessary sequencing reads. Furthermore, to ensure we provide panels with balanced reads across a number of different biological sample types, we test our cocktails with multiple donors, on multiple cell types, and under different stimulation conditions. Together our optimized antibody cocktail formulation process allows us to provide ready-to-use, pre-titrated, balanced antibody panels with unmatched performance.
TotalSeq™ Oligonucleotide-Antibody Cocktails
TotalSeq™ antibody cocktails are provided in convenient, single-use tubes and offer reduced costs when compared to buying individual antibodies. They can also minimize variability between different labs or across longitudinal studies. We now offer 3 format-specific Human Universal Cocktails, featuring over 130 antibodies each. We also offer a smaller panel, the Human TBNK Cocktail, containing 9 antibodies for the identification of principal lineage immune cell types. Lastly, the most recent addition to our cocktail portfolio is the TotalSeq™-D Human Heme Oncology cocktail, developed specifically for single-cell DNA and protein studies on the Mission Bio Tapestri platform.
Have confidence in your antibody panels and try one of our comprehensively validated TotalSeq™ cocktails in your next CITE-seq experiment.
 Login / Register
Login / Register 






Follow Us